Overview
GATK-SV consists of multiple modules that need to be executed in a specific order. For joint calling, each module must be executed individually in sequence. While the single-sample mode invokes many of these modules, it is implemented as a single runnable workflow.
The following diagram illustrates the overall module ordering:
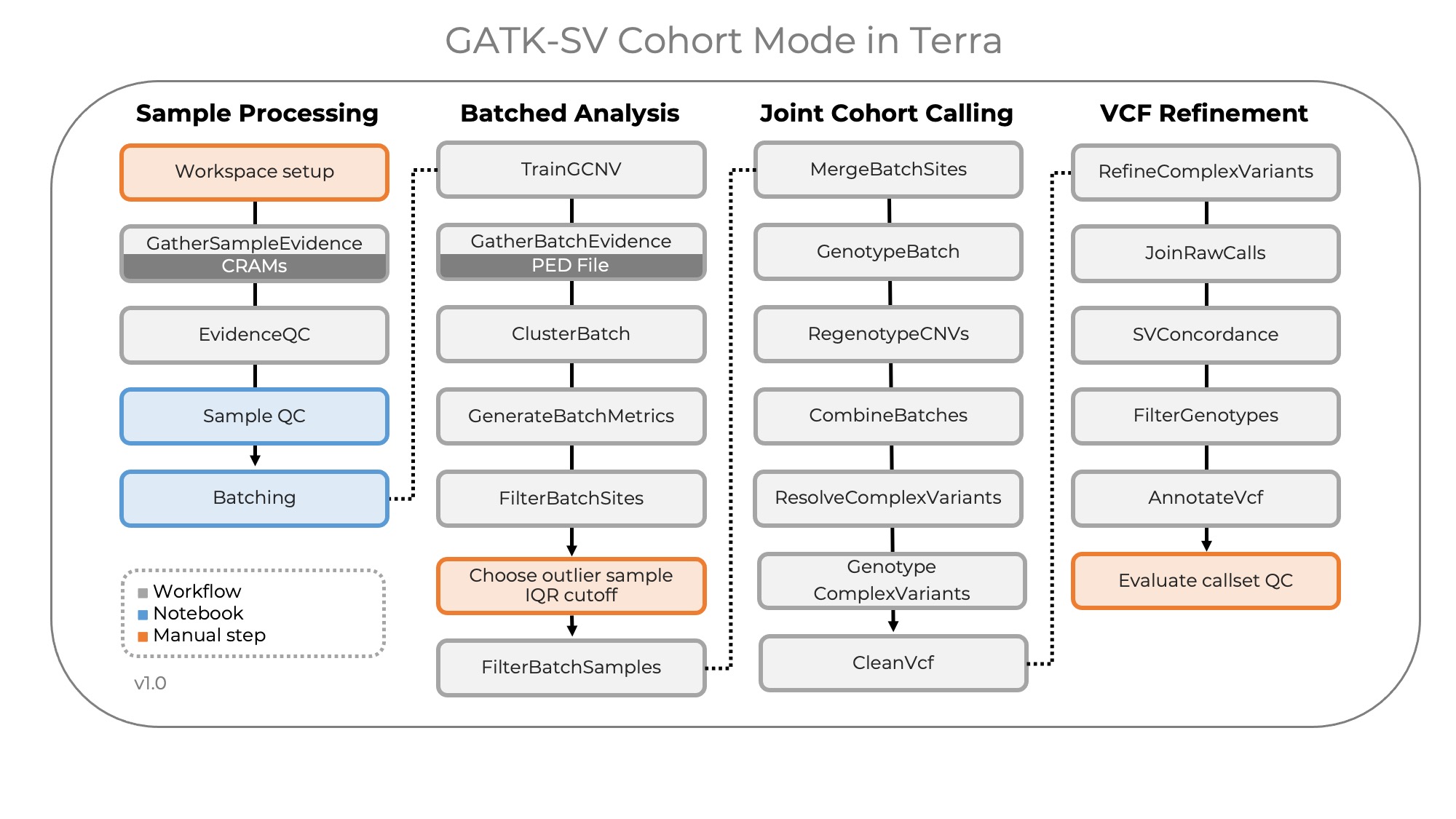
Each module is implemented in the Workflow Description Language (WDL). The Terra workspaces come pre-configured with default values for all required parameters and set up to run the pipeline for most use cases.
The following sections supplement the Terra workspaces with documentation for each WDL, including an overview of its function, key input parameters, and outputs. Not all parameters are documented here, as some WDLs contain dozens of inputs. Descriptions of some common inputs can be found in the Resource files and Runtime attributes sections. Users are encouraged to refer to the WDL source code for additional clarification.
For details on running GATK-SV on Terra, refer to the Execution section.