Quick start
The commands below will annotate some example MAS-seq 15-element array reads from a synthetic set of RNA isoforms (Lexogen SIRV set 4), segment arrays into individual array elements (e.g. generate a separate read per array element), extract cDNA sequences, and align them to the SIRV reference with pbmm2. Assuming your Longbow installation is at $LONGBOW_DIR, run the following two commands:
$ longbow annotate $LONGBOW_DIR/tests/test_data/mas15_test_input.bam | longbow segment | longbow extract -o extracted.bam
[INFO 2021-08-08 22:35:07 extract] Invoked via: longbow extract -o extracted.bam
[INFO 2021-08-08 22:35:07 segment] Invoked via: longbow segment
[INFO 2021-08-08 22:35:07 annotate] Invoked via: longbow annotate tests/test_data/mas15_test_input.bam
[INFO 2021-08-08 22:35:07 extract] Writing extracted read segments to: extracted.bam
[INFO 2021-08-08 22:35:07 segment] Running with 11 worker subprocess(es)
[INFO 2021-08-08 22:35:07 extract] Extracting `random` segments between 10x_Adapter and Poly_A.
[INFO 2021-08-08 22:35:07 segment] Using bounded region splitting mode.
[INFO 2021-08-08 22:35:07 annotate] Running with 11 worker subprocess(es)
[INFO 2021-08-08 22:35:07 extract] Ignoring the first 26 bases from extracted read segments.
[INFO 2021-08-08 22:35:07 annotate] Using The standard MAS-seq 15 array element model.
[INFO 2021-08-08 22:35:07 extract] Including 2 flanking bases.
[INFO 2021-08-08 22:35:09 annotate] Annotating 8 reads
[INFO 2021-08-08 22:35:10 segment] Using The standard MAS-seq 15 array element model.
Progress: 100%|██████████████████████████████████████████████████████████████████████████████| 8/8 [00:01<00:00, 5.60 read/s]
Progress: 100%|██████████████████████████████████████████████████████████████████████████████| 8/8 [00:01<00:00, 6.01 read/s]
[INFO 2021-08-08 22:35:10 annotate] Annotated 8 reads with 572 total sections.
[INFO 2021-08-08 22:35:10 annotate] Done. Elapsed time: 3.78s. Overall processing rate: 2.12 reads/s.
[INFO 2021-08-08 22:35:13 segment] Using The standard MAS-seq 15 array element model.
[INFO 2021-08-08 22:35:15 segment] Segmented 8 reads with 108 total segments.
[INFO 2021-08-08 22:35:15 segment] Done. Elapsed time: 8.62s.
[INFO 2021-08-08 22:35:15 extract] Done. Elapsed time: 8.77s.
[INFO 2021-08-08 22:35:15 extract] Total # Reads Processed: 108
[INFO 2021-08-08 22:35:15 extract] # Reads Containing Extracted Segments: 108 (100.00%)
[INFO 2021-08-08 22:35:15 extract] Total # Segments Extracted: 108
[INFO 2021-08-08 22:35:15 extract] Total # Segments Skipped: 0
[INFO 2021-08-08 22:35:15 extract] # Segments extracted per read: 1.00
$ pbmm2 align --preset ISOSEQ --sample SIRVTest --sort tests/test_data/resources/SIRV_Library.fasta extracted.bam extracted.aligned.bam
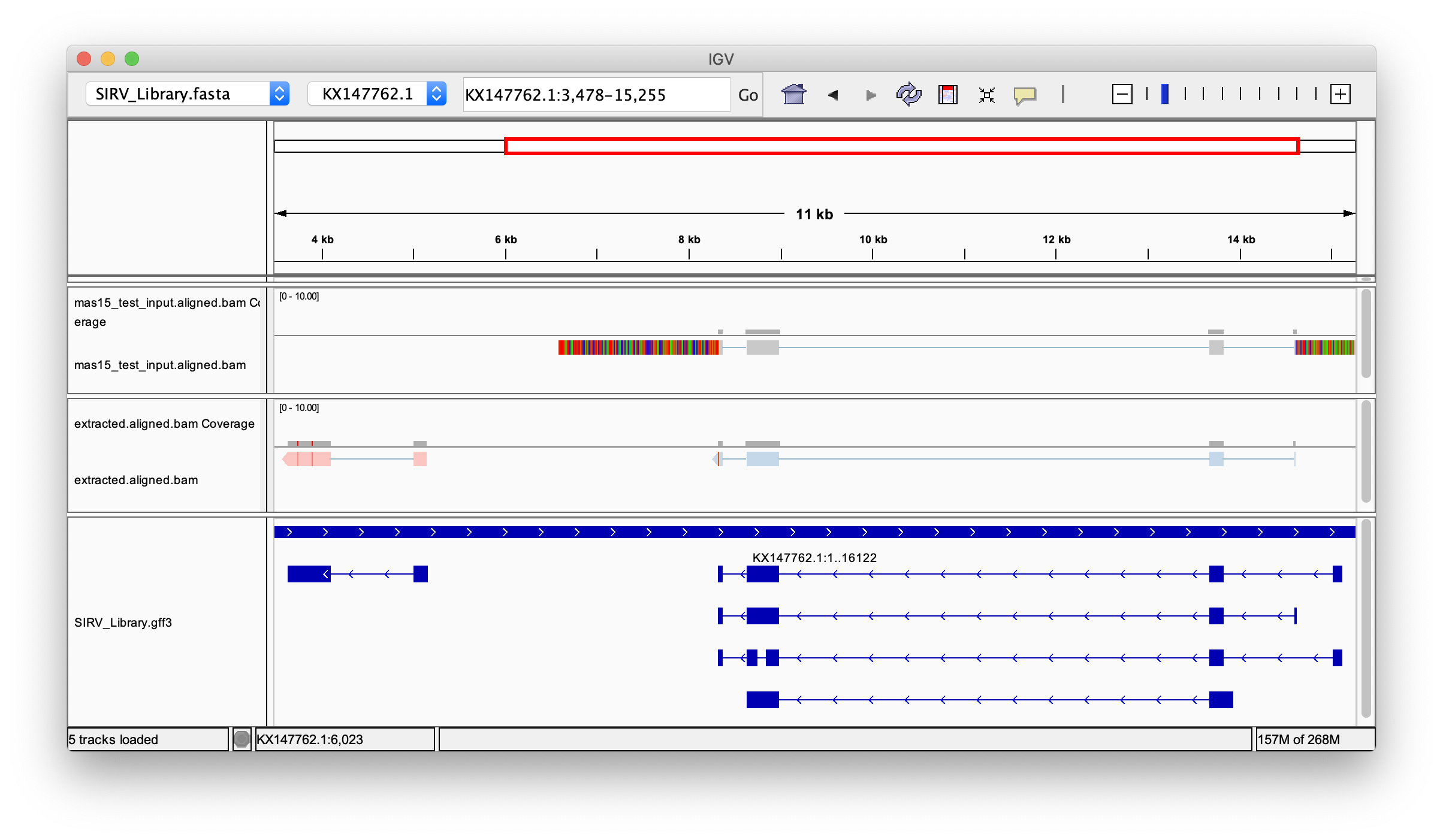
Below, we examine the data in IGV before (top track) and after Longbow processing (middle track) over two of the SIRV isoforms (bottom track; locus: KX147762.1:3478-15255). Notice the soft-clipped bases (the myriad reference-mismatching bases at the 5’ and 3’ ends of the read) in the data before Longbow processing. These represent the other concatenated transcripts that have not been separated from each other yet. After segmentation by Longbow, each read now appropriately represents an individual transcript isoform sequence and is ready for downstream analysis.