Inspect
Description
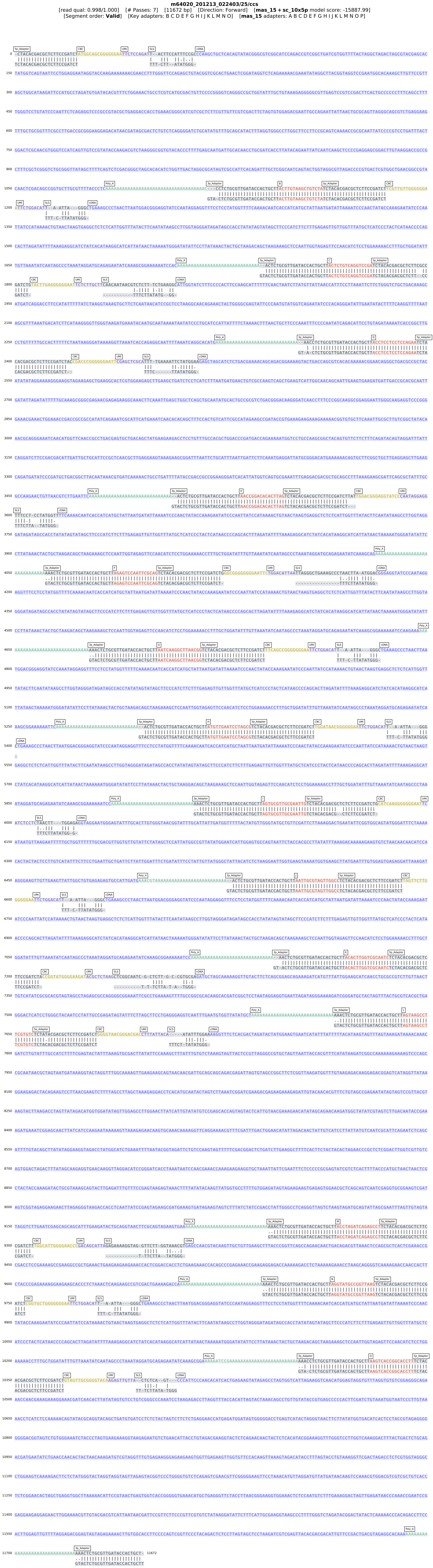
It can occasionally be useful to inspect the classifications made by Longbow’s hidden Markov model. To simplify this process, Longbow provides the inspect command, which can take existing annotations (or redo the annotation from scratch) and display the full sequence of the read with the annotated adapters color-coded appropriately.
Output in .png and .pdf formats are supported via the --file-format argument. Choose ‘png’ if you want a rasterized image for a quick look at data. For a vector image that also has the added capability of highlighting and copying read subsequences, choose the ‘pdf’ output.
If a .pbi file for the input .bam file is available, then specific reads can be fetched very quickly without iterating over the entire file. If the .pbi file is not available, the BAM file will be scanned linearly until the requested reads are found. The .pbi file is therefore highly recommended and can be easily generated using the pbindex tool, installable via conda install pbbam.
Command help
$ longbow inspect --help
Usage: longbow inspect [OPTIONS] INPUT_BAM
Inspect the classification results on specified reads.
Options:
-v, --verbosity LVL Either CRITICAL, ERROR, WARNING, INFO or DEBUG
-r, --read-names TEXT read names (or file(s) of read names) to
inspect
-p, --pbi PATH BAM .pbi index file
-f, --file-format [png|pdf] Image file format [default: pdf]
-o, --outdir PATH Output directory [default: .]
-m, --model TEXT The model to use for annotation. If the given
value is a pre-configured model name, then
that model will be used. Otherwise, the given
value will be treated as a file name and
Longbow will attempt to read in the file and
create a LibraryModel from it. Longbow will
assume the contents are the configuration of a
LibraryModel as per LibraryModel.to_json().
--seg-score Display alignment score for annotated
segments. (--quick mode only) [default:
False]
--max-length INTEGER Maximum length of a read to process. Reads
beyond this length will not be annotated. If
the input file has already been annotated,
this parameter is ignored. [default: 30000]
--min-rq FLOAT Minimum ccs-determined read quality for a read
to be annotated. CCS read quality range is
[-1,1]. If the input file has already been
annotated, this parameter is ignored.
[default: -2]
-q, --quick Create quick (simplified) inspection figures.
[default: False]
-a, --annotated-bam FILENAME Store annotations from a downstream BAM file
so they can be displayed on reads from
previous processing steps.
--help Show this message and exit.
Example
$ longbow inspect -m mas_15+sc_10x5p -r m64020_201213_022403/25/ccs -o images tests/test_data/mas15_test_input.bam
[INFO 2022-11-25 20:29:36 inspect] Invoked via: longbow inspect -m mas_15+sc_10x5p -r m64020_201213_022403/25/ccs -o images tests/test_data/mas15_test_input.bam
[INFO 2022-11-25 20:29:37 inspect] Using mas_15+sc_10x5p: 15-element MAS-ISO-seq array, single-cell 10x 5' kit
[INFO 2022-11-25 20:29:37 inspect] Figure drawing mode: extended
[INFO 2022-11-25 20:29:37 inspect] No .pbi file available. Inspecting whole input bam file until we find specified reads.
[INFO 2022-11-25 20:29:37 inspect] Drawing read 'm64020_201213_022403/25/ccs' to 'images/m64020_201213_022403_25_ccs.pdf'
[INFO 2022-11-25 20:31:18 inspect] Done. Elapsed time: 101.38s.
An example screenshot from the longbow inspect command can be found below. Note that for visual clarity, random model sections are drawn as gray read sections. Only adapter sequences and poly-A tails are labeled and color-coded.