15 Pseudotime Cell Trajectories
- Comparison: Cell Trajectories
- Diffusion maps for high-dimensional single-cellanalysis
- Diffusion pseudotime
- Slingshot Paper
- Optimal Transport
- RNA Velocity
15.1 Google Slides
15.2 Comparison Abstract
Using single-cell -omics data, it is now possible to computationally order cells along trajectories, allowing the unbiased study of cellular dynamic processes. Since 2014, more than 50 trajectory inference methods have been developed, each with its own set of methodological characteristics. As a result, choosing a method to infer trajectories is often challenging, since a comprehensive assessment of the performance and robustness of each method is still lacking. In order to facilitate the comparison of the results of these methods to each other and to a gold standard, we developed a global framework to benchmark trajectory inference tools. Using this framework, we compared the trajectories from a total of 29 trajectory inference methods, on a large collection of real and synthetic datasets. We evaluate methods using several metrics, including accuracy of the inferred ordering, correctness of the network topology, code quality and user friendliness. We found that some methods, including Slingshot (Street et al. 2018), TSCAN (Z. Ji and Ji 2016) and Monocle DDRTree (Trapnell et al. 2014), clearly outperform other methods, although their performance depended on the type of trajectory present in the data. Based on our benchmarking results, we therefore developed a set of guidelines for method users. However, our analysis also indicated that there is still a lot of room for improvement, especially for methods detecting complex trajectory topologies. Our evaluation pipeline can therefore be used to spearhead the development of new scalable and more accurate methods, and is available at github.com/dynverse/dynverse. (Saelens et al. 2018)
(Haghverdi, Buettner, and Theis 2015) (Haghverdi et al. 2016) (Street et al. 2018) (Schiebinger et al. 2019) (La Manno et al. 2018)
knitr::include_graphics("images/PseudotimeComparisonGuidelines.png")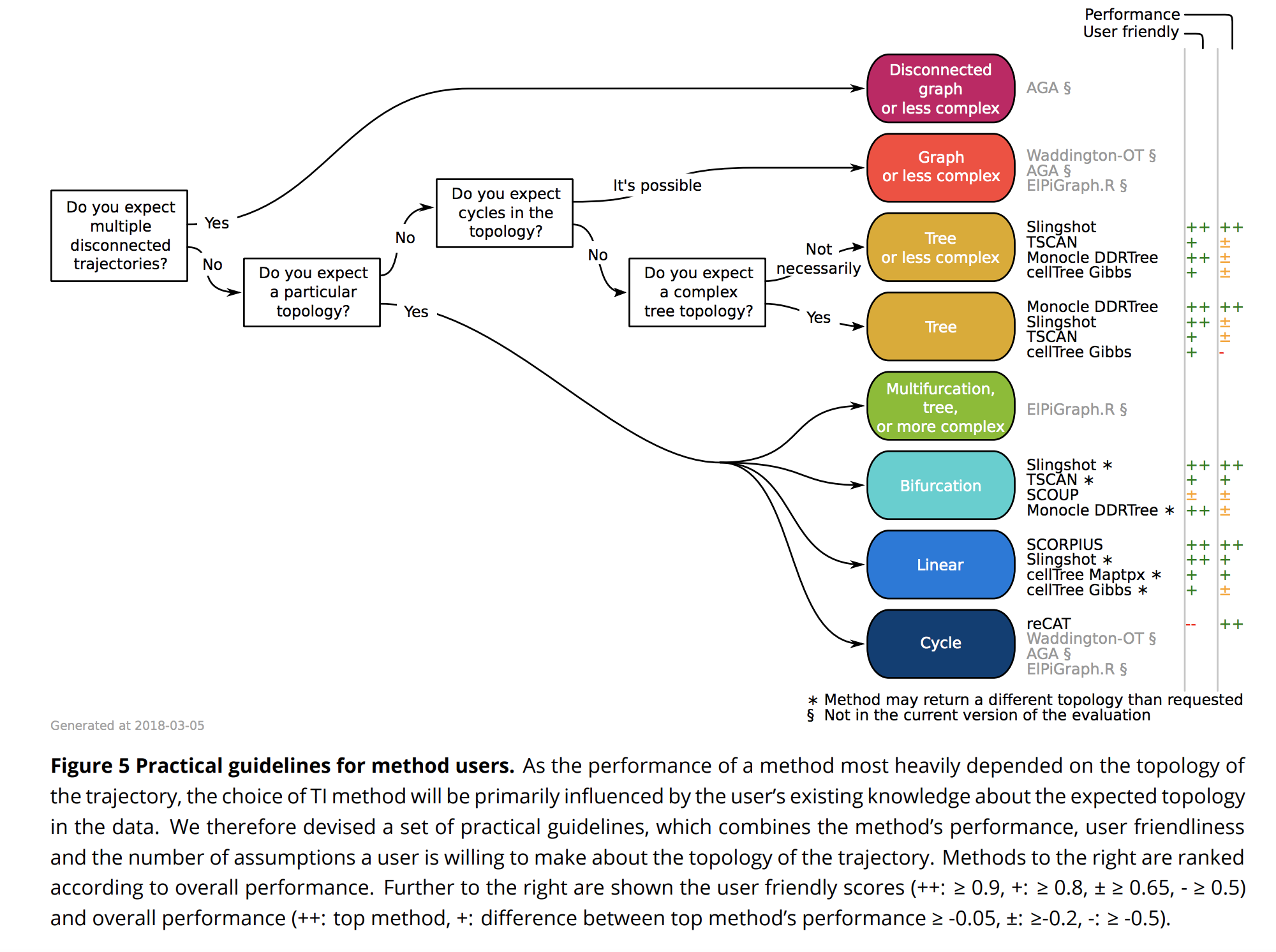
References
Street, Kelly, Davide Risso, Russell B Fletcher, Diya Das, John Ngai, Nir Yosef, Elizabeth Purdom, and Sandrine Dudoit. 2018. “Slingshot: Cell Lineage and Pseudotime Inference for Single-Cell Transcriptomics.” BMC Genomics 19 (1). BioMed Central: 477.
Ji, Zhicheng, and Hongkai Ji. 2016. “TSCAN: Pseudo-Time Reconstruction and Evaluation in Single-Cell Rna-Seq Analysis.” Nucleic Acids Research 44 (13). Oxford University Press: e117–e117.
Trapnell, Cole, Davide Cacchiarelli, Jonna Grimsby, Prapti Pokharel, Shuqiang Li, Michael Morse, Niall J Lennon, Kenneth J Livak, Tarjei S Mikkelsen, and John L Rinn. 2014. “The Dynamics and Regulators of Cell Fate Decisions Are Revealed by Pseudotemporal Ordering of Single Cells.” Nature Biotechnology 32 (4). Nature Publishing Group: 381.
Saelens, Wouter, Robrecht Cannoodt, Helena Todorov, and Yvan Saeys. 2018. “A Comparison of Single-Cell Trajectory Inference Methods: Towards More Accurate and Robust Tools.” bioRxiv. Cold Spring Harbor Laboratory, 276907.
Haghverdi, Laleh, Florian Buettner, and Fabian J Theis. 2015. “Diffusion Maps for High-Dimensional Single-Cell Analysis of Differentiation Data.” Bioinformatics 31 (18). Oxford University Press: 2989–98.
Haghverdi, Laleh, Maren Buettner, F Alexander Wolf, Florian Buettner, and Fabian J Theis. 2016. “Diffusion Pseudotime Robustly Reconstructs Lineage Branching.” Nature Methods 13 (10). Nature Publishing Group: 845.
Schiebinger, Geoffrey, Jian Shu, Marcin Tabaka, Brian Cleary, Vidya Subramanian, Aryeh Solomon, Joshua Gould, et al. 2019. “Optimal-Transport Analysis of Single-Cell Gene Expression Identifies Developmental Trajectories in Reprogramming.” Cell. Elsevier.
La Manno, Gioele, Ruslan Soldatov, Amit Zeisel, Emelie Braun, Hannah Hochgerner, Viktor Petukhov, Katja Lidschreiber, et al. 2018. “RNA Velocity of Single Cells.” Nature 560 (7719). Nature Publishing Group: 494.