A whistlestop tour of the book contents
It can be hard to get a sense of whether a book like ours is right for you just based on the Amazon listing (which we don’t have direct control over as authors). To address that problem, I figured I’d write a short series of blog posts dedicated to providing insight into “What’s in the can?”, ie what our book is about, and what it’s like to work through, to supplement the information provided in the listing.
In this first installment, I adapted a Twitter thread I had posted a while back that people seemed to find helpful, consisting of one tweet per chapter, each providing a brief and informal description of the chapter’s topic and goals, with a sampling of illustrations from that chapter. The format and Twitter’s brutally low character limit forced me to be very concise, and really drill down to the essentials. The result emphasizes the progression from one topic to the next, and reflects our vision of the book as an educational journey rather than a collection of facts.
Here we go.
1. Introduction
Why you should care about the cloud, and how bioinformatics / life sciences research benefits from moving to a cloud-based ecosystem for data sharing and analysis. No, the cloud environment is not perfect; yes, it really is a game changer.
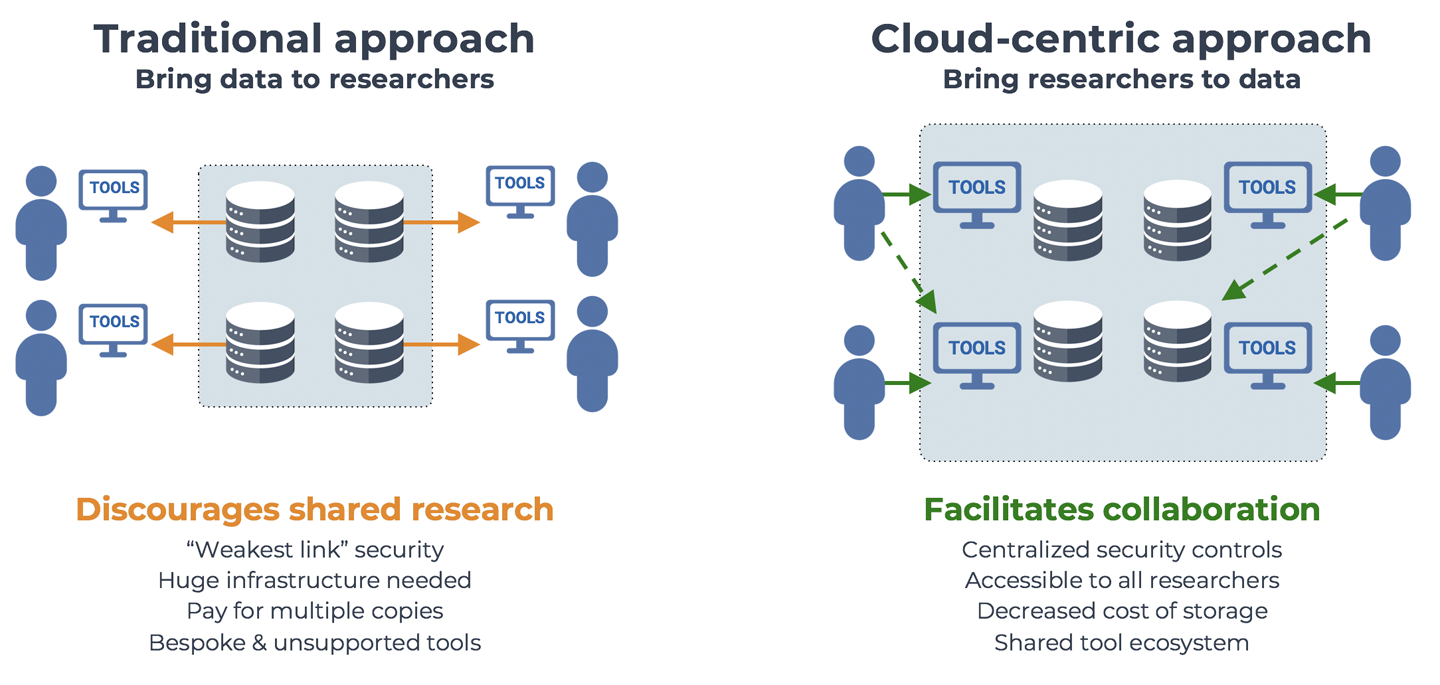
2. Genomics in a Nutshell
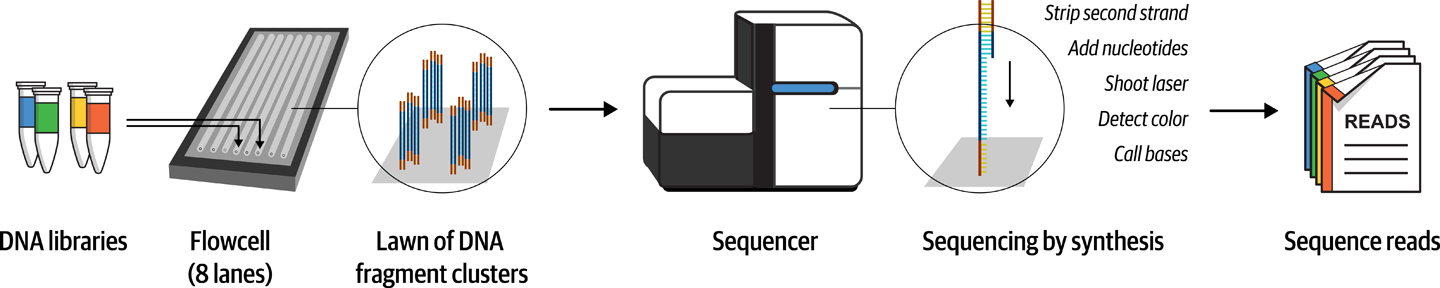
A primer for newcomers to the field of genomics, covering foundational terms and concepts such as genes, DNA and genomic variation, plus the technical basics of sequencing and handling genomic data.
—
3. Computing Technology Basics for Life Scientists
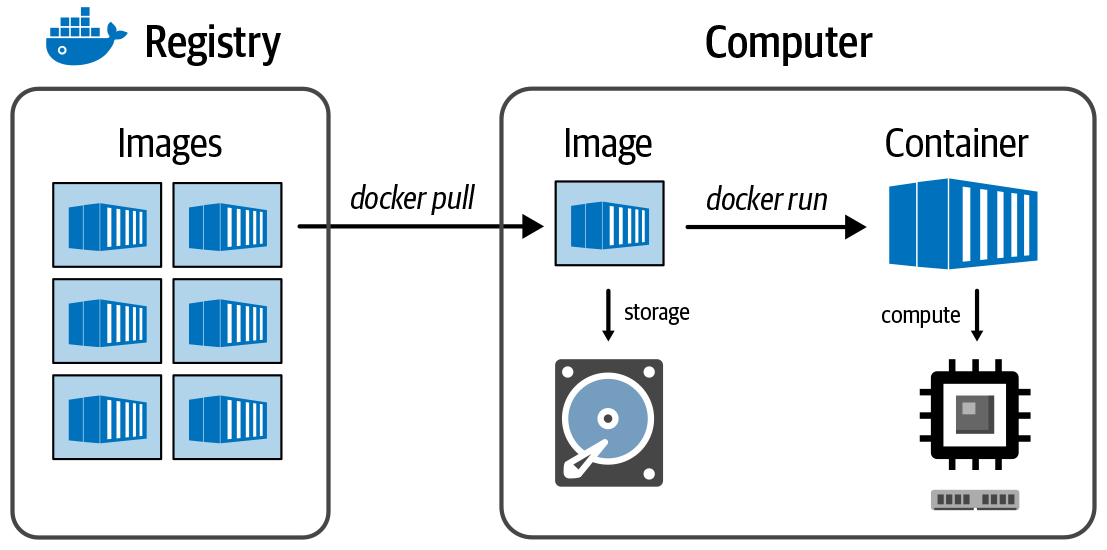
CPU, GPU, TPU, FPGA, OMG GTFO – no really, just some basic hardware terminology, plus an introduction to key concepts like parallelism, pipelining, containers and virtual machines in fairly plain language.
4. First Steps in the Cloud
Finally we get to do some hands-on work (on Google Cloud). Set up an account, get free credits, practice managing data in storage buckets and interacting with a Docker container, get a nice custom VM set up to do some genomics.
—
5. First Steps with GATK
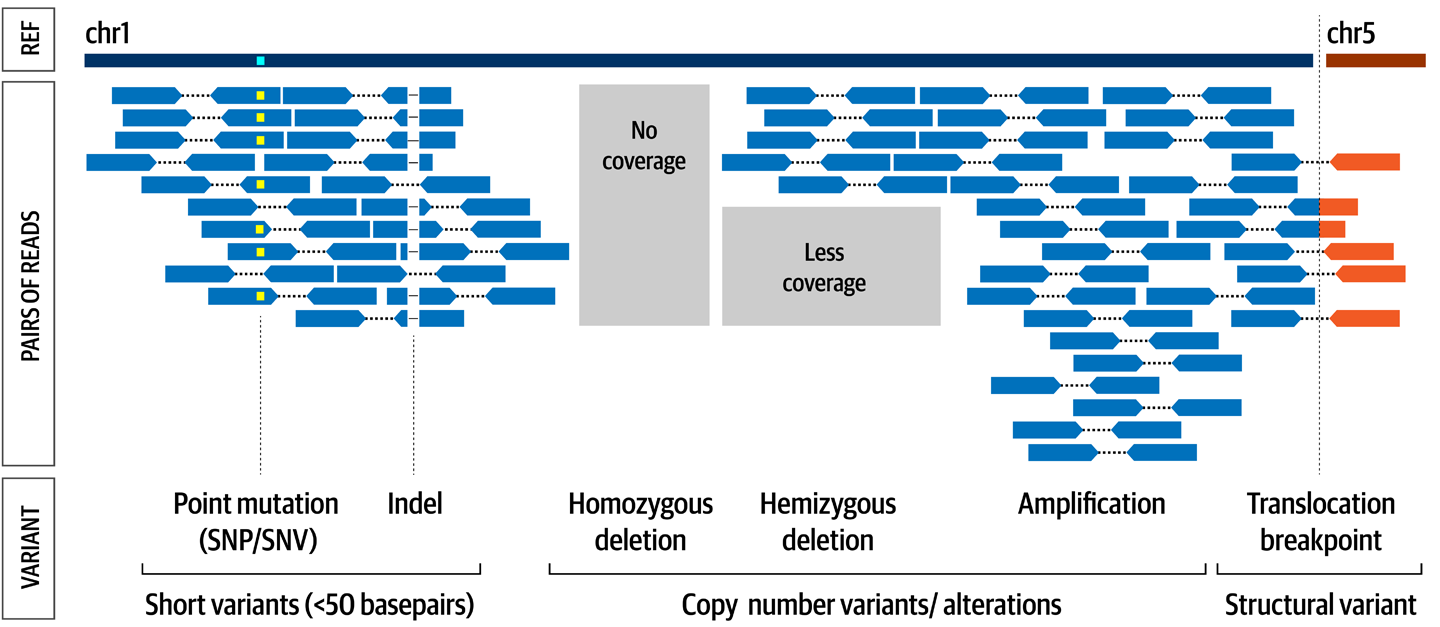
Let’s meet the workhorse of genomics! We start with a general overview, requirements, command line syntax, the usual – then dive into calling variants with HaplotypeCaller, plus some visual troubleshooting and variant filtering concepts.
—
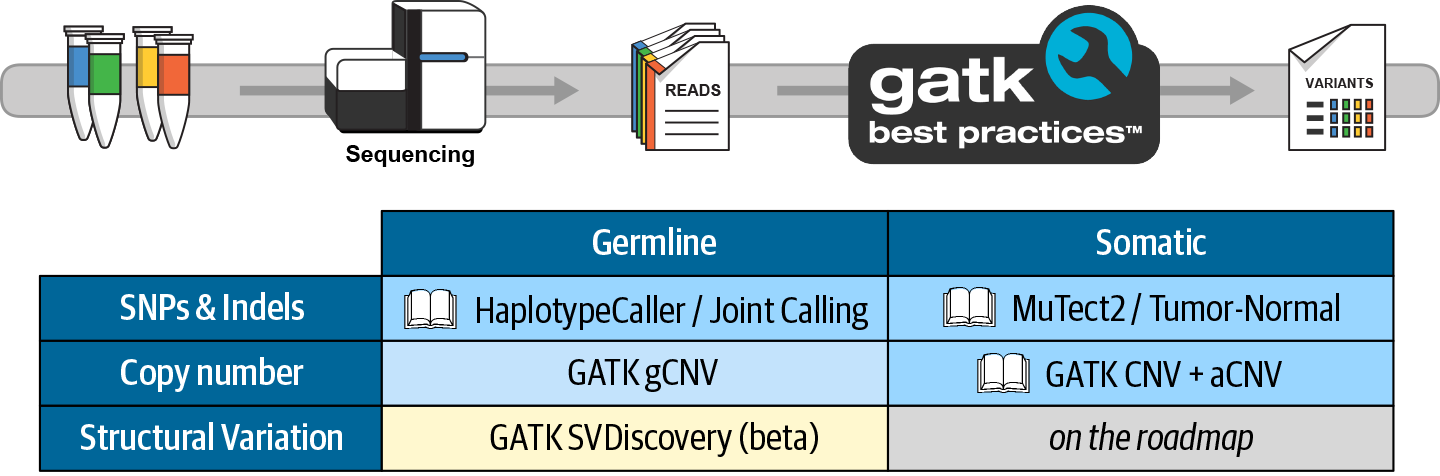
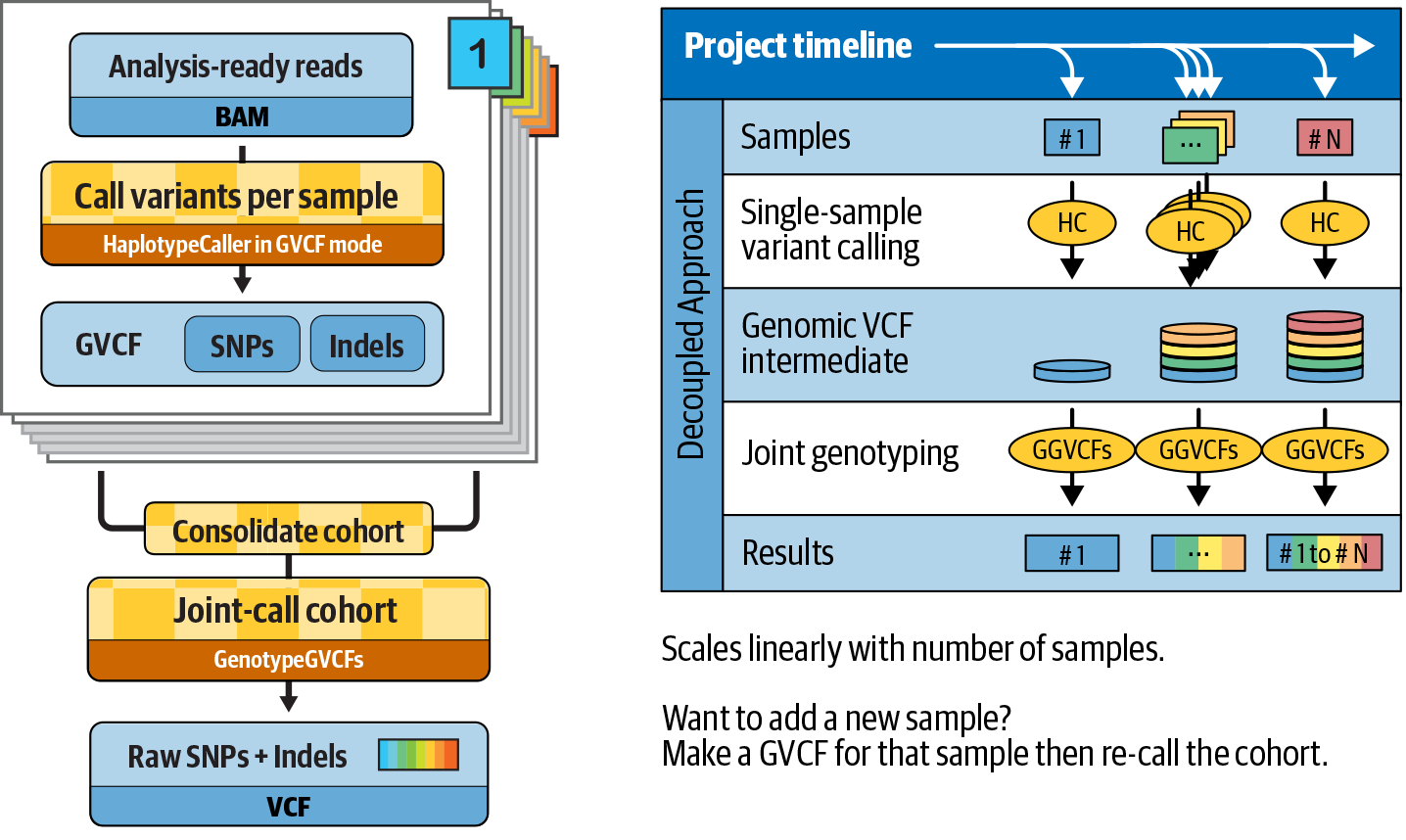
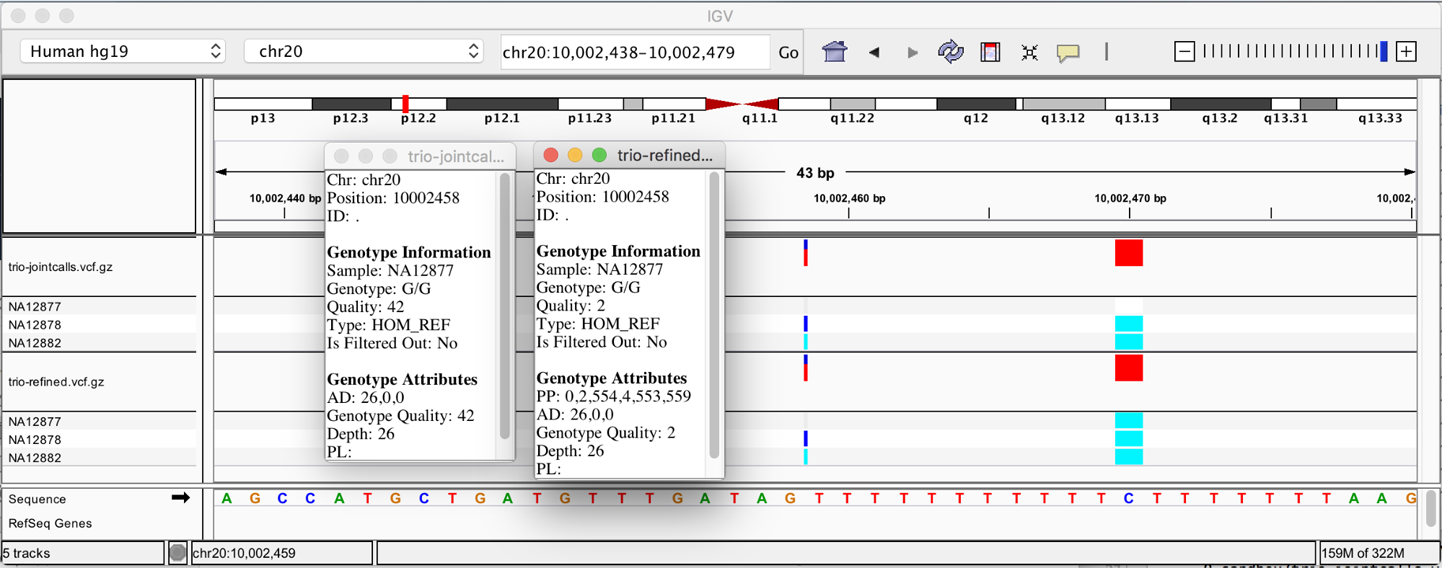
6. GATK Best Practices for Germline Short Variant Discovery
Step by step examination of what may be the most commonly run genomics pipeline in the world, with highlights on joint calling for populations and deep learning for single-sample analysis.
—
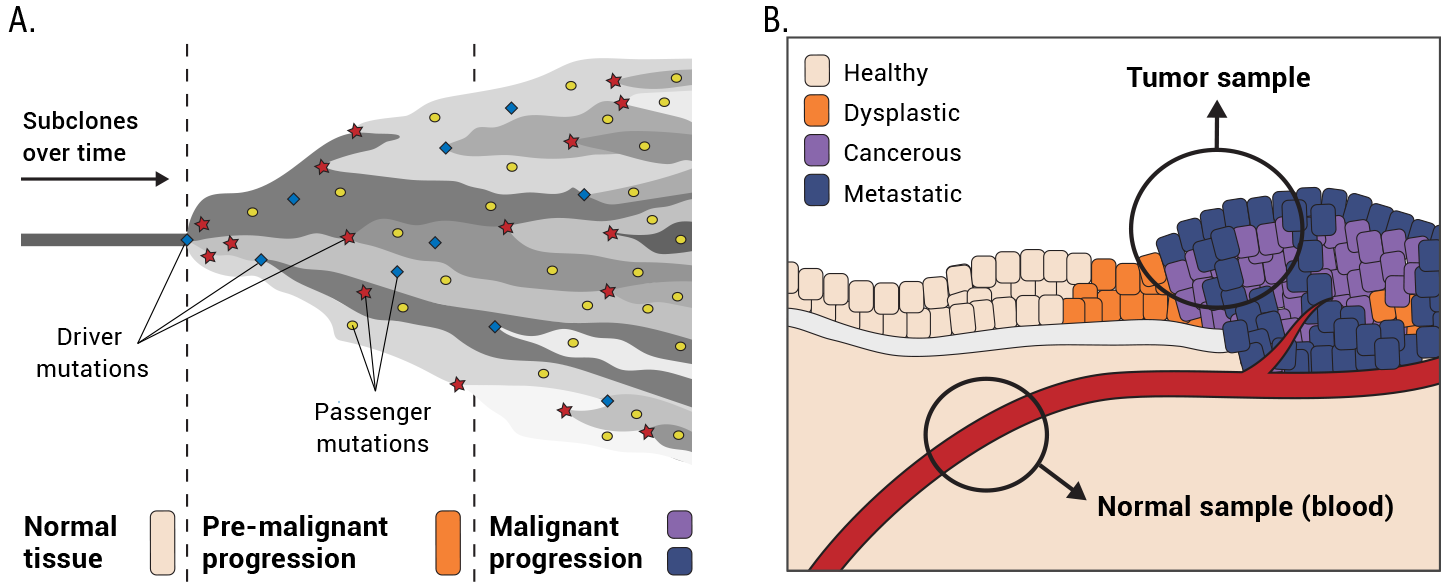
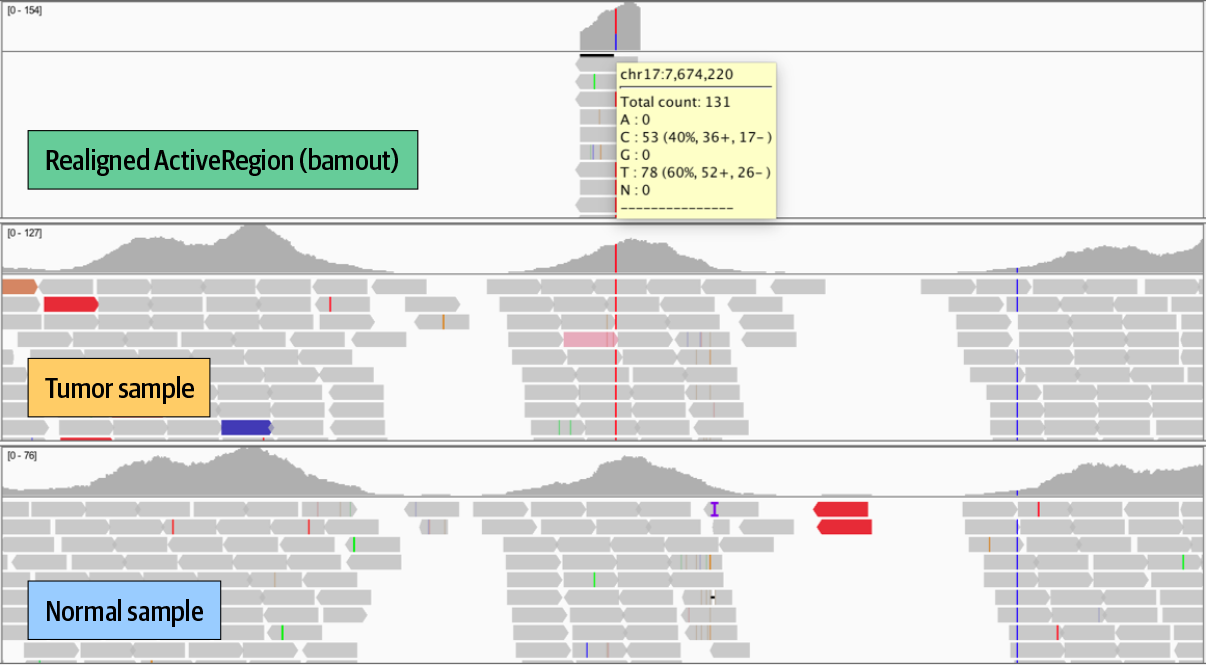
7. GATK Best Practices for Somatic Variant Discovery
Switching gears to cancer genomics with a rundown of how somatic calling is different; step by step through the pipelines for somatic short variants (Mutect2) and copy number alterations.
—
8. Automating Analysis Execution with Workflows
Halfway point; we pivot to the challenges of automating and scaling up these analyses, introducing the Cromwell workflow system and the portable Workflow Description Language (WDL).
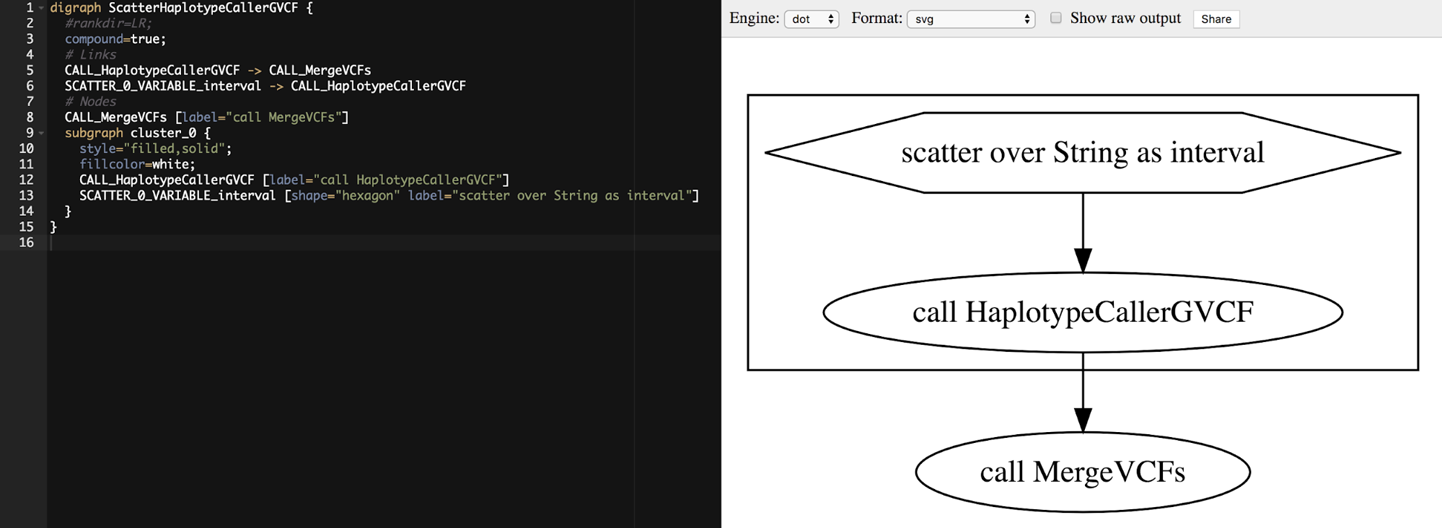
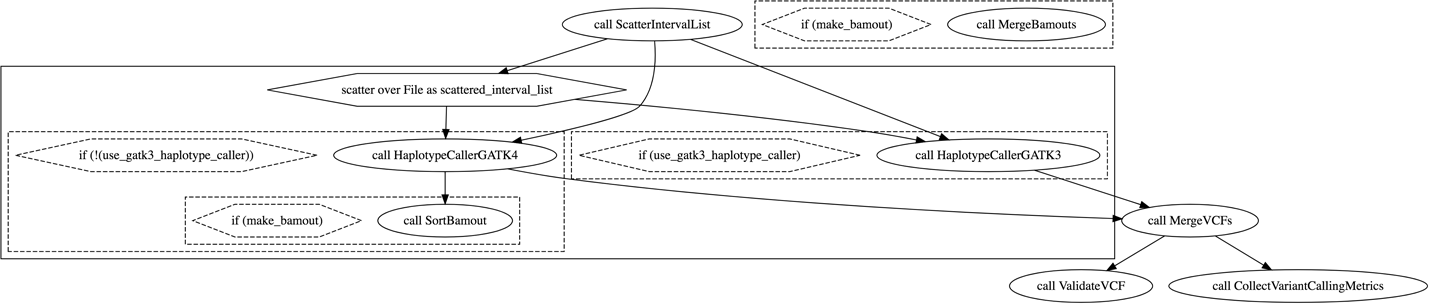
9. Deciphering Real Genomics Workflows
We pretend to stumble across 2 mystery workflows, go through a systematic process of investigating their content to understand what they do and how they do it, learning useful WDL features along the way.
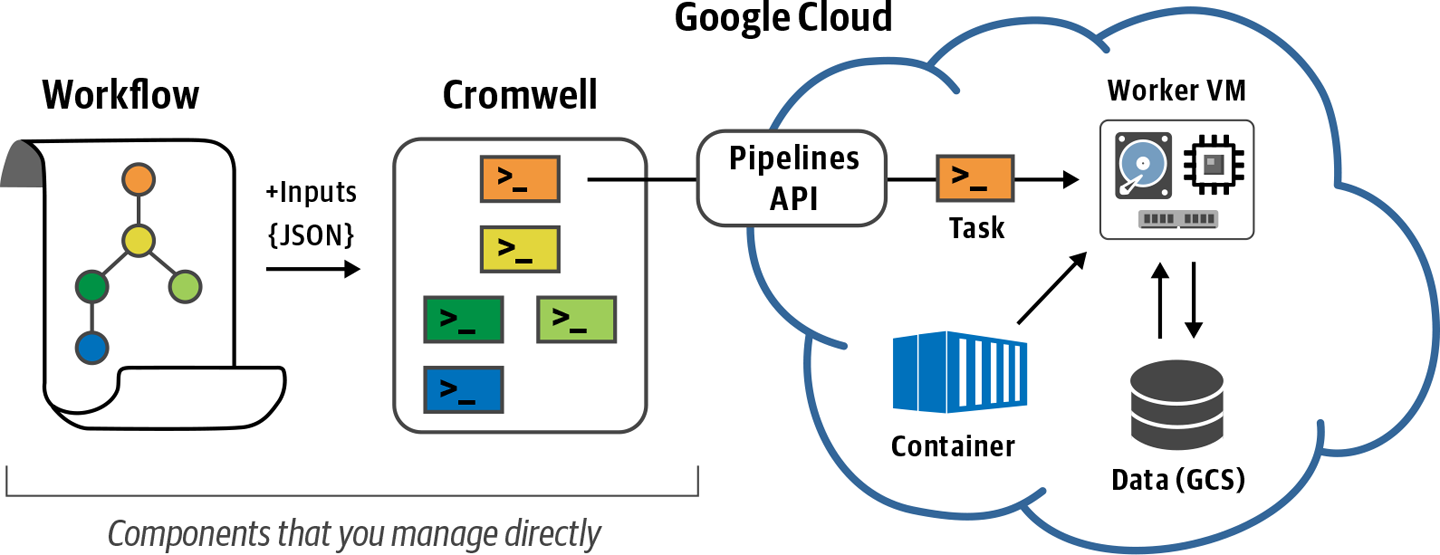
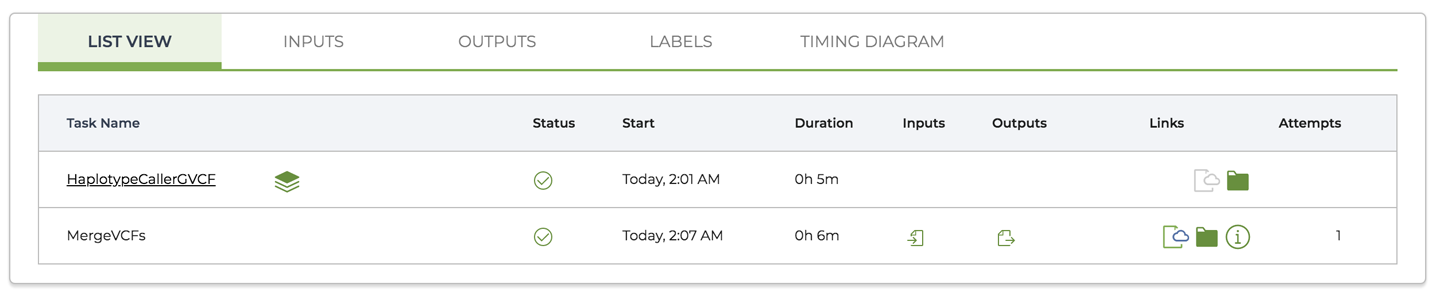
10. Running Single Workflows at Scale with Pipelines API
So far we’ve been running everything on our little custom VM. Now it’s time to unleash the full power of the cloud by dispatching workflow tasks to multiple machines – with surprisingly little effort.
—
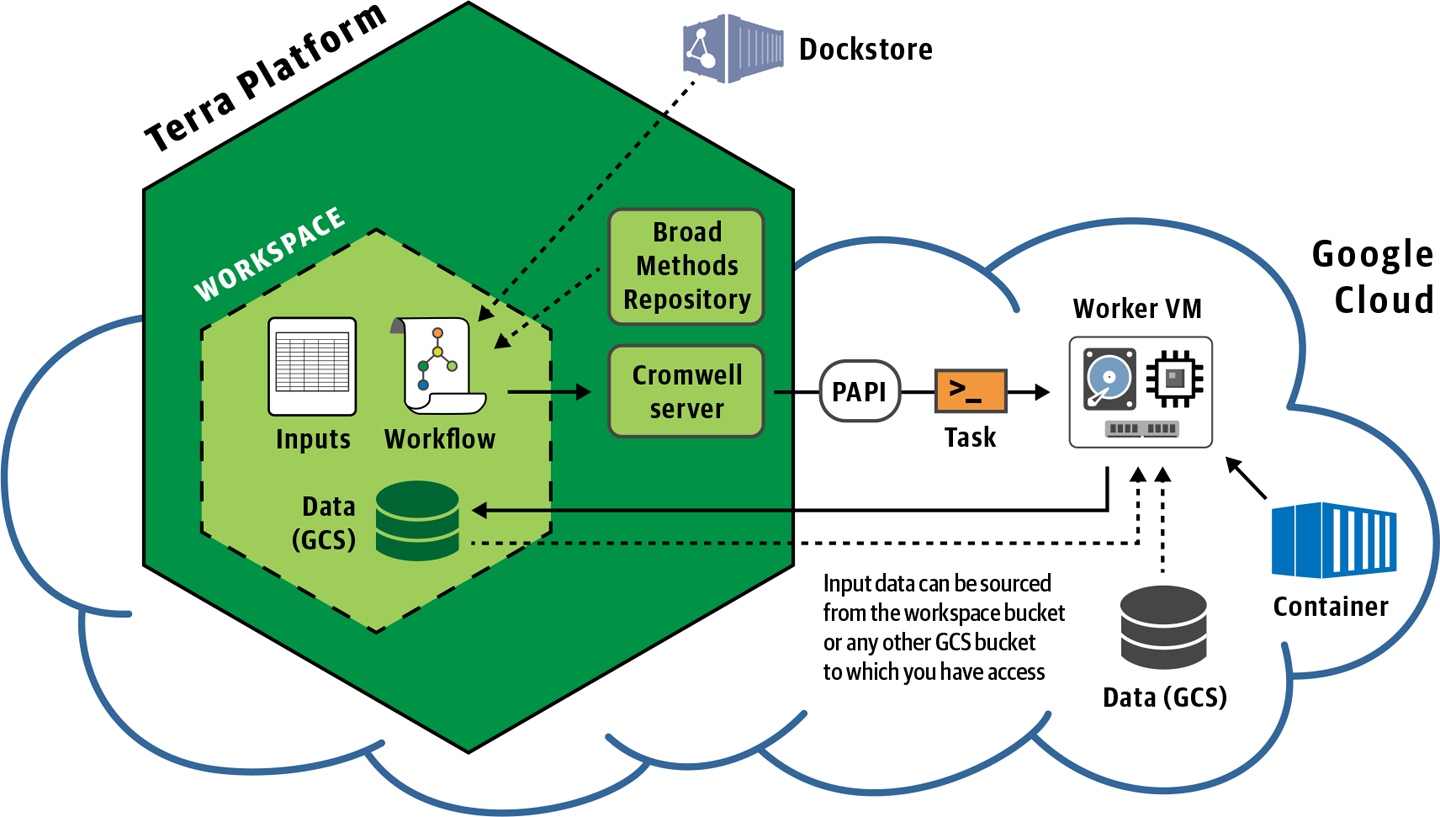
11. Running Many Workflows Conveniently in Terra
Now we’re scaling up to arbitrary numbers of samples, using the managed Cromwell server in Terra, an open platform for secure data access and analysis.
—
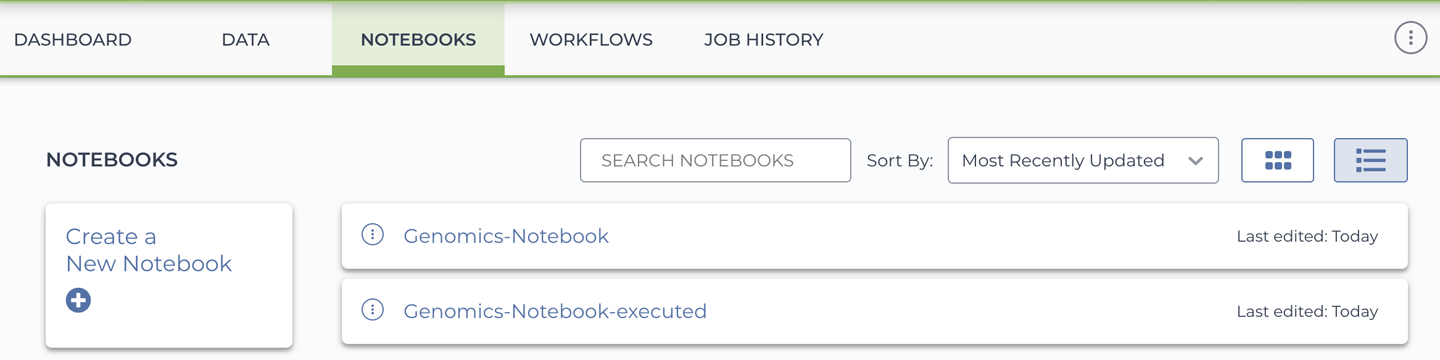
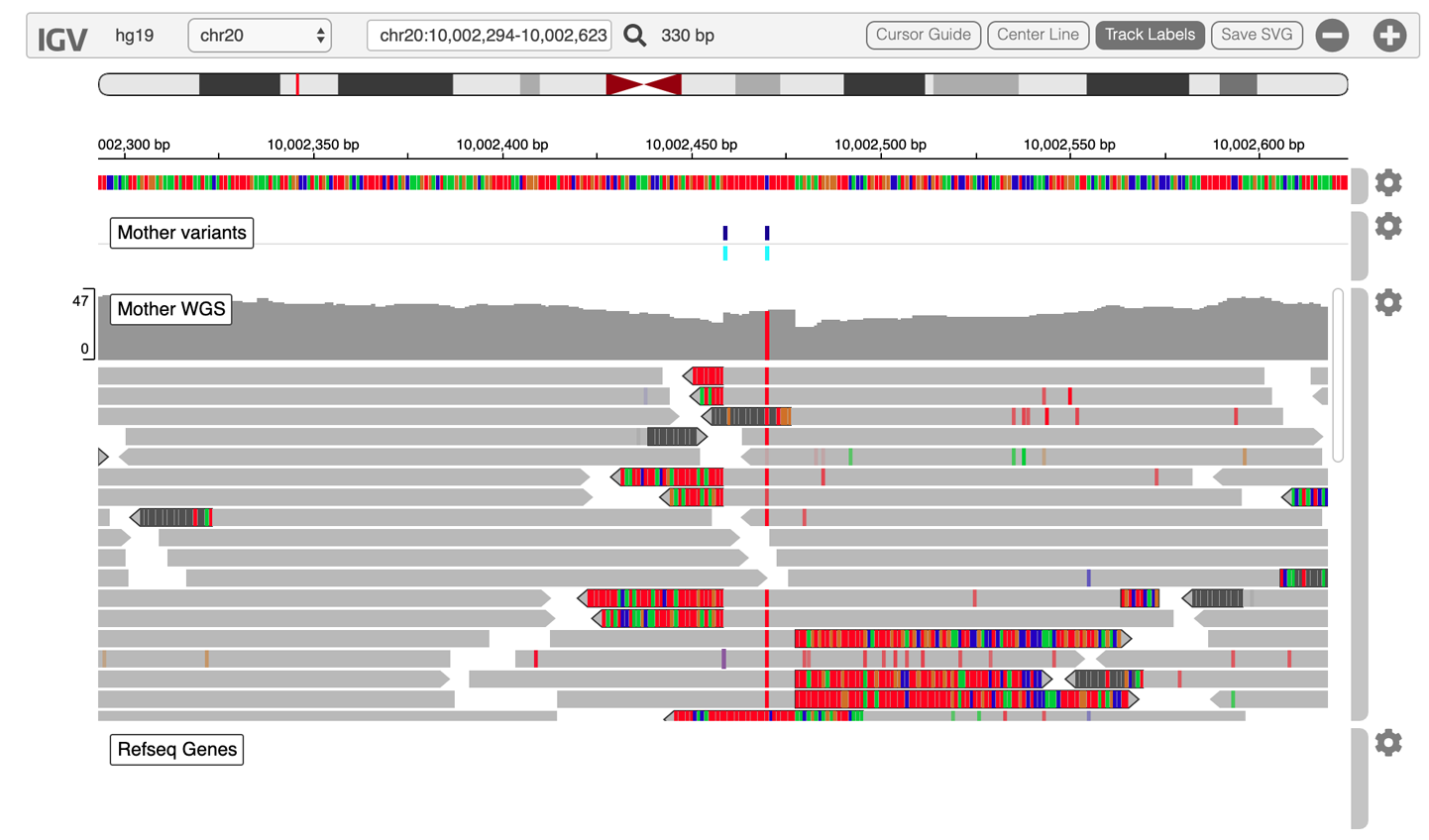
12. Interactive Analysis in Jupyter Notebook
Circling back to the GATK work from earlier chapters, we examine what that would all look like done in Jupyter Notebooks instead of the terminal shell. Between embedded IGV and ggplots galore, it looks good!
—

—
13. Assembling Your Own Workspace in Terra
Crossing the bridge from canned examples to importing your own data and methods into Terra in a few different scenarios. Draws on other services in the ecosystem including workflow and data repositories.
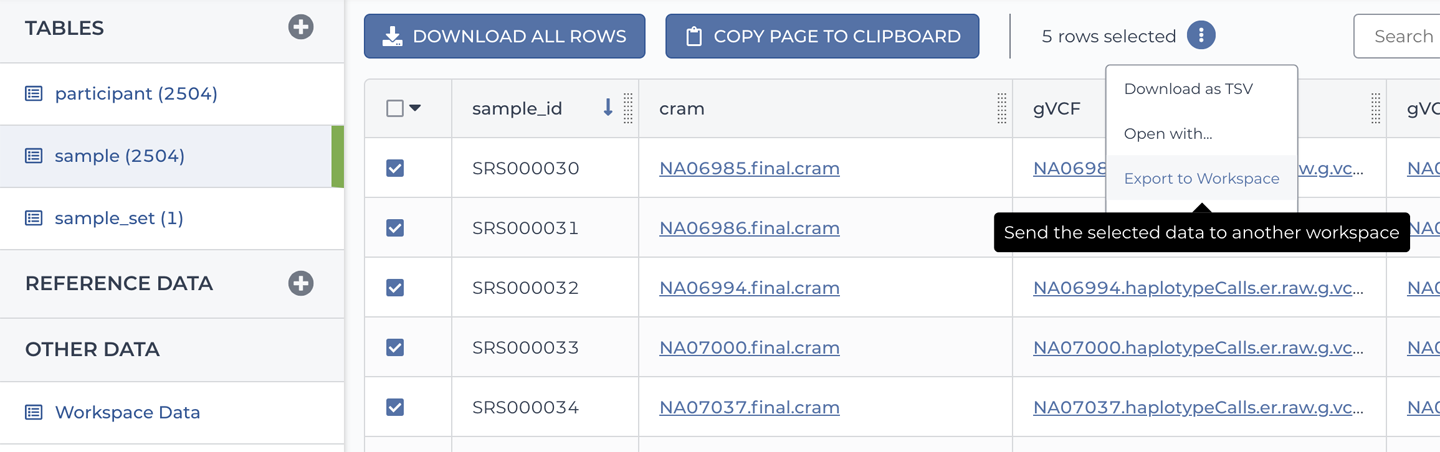
—
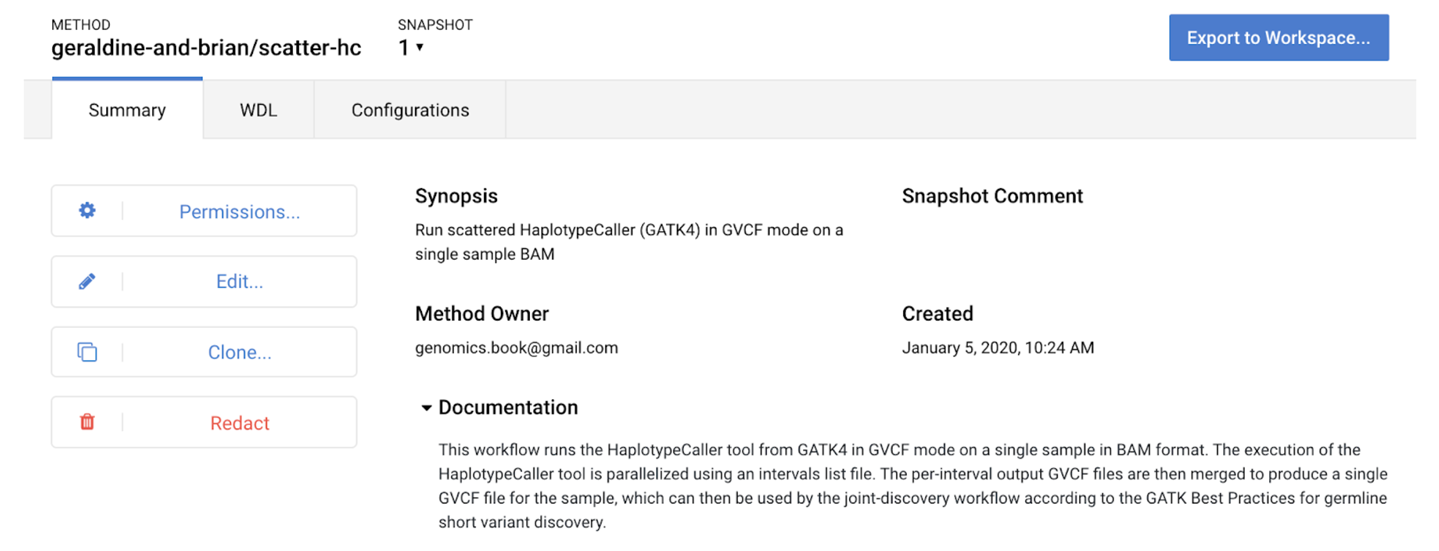
14. Making a Fully Reproducible Paper
Capstone case study on computational reproducibility involving synthetic data creation, GATK, downstream analysis and real biological findings by Dr. Matthieu Miossec et al.
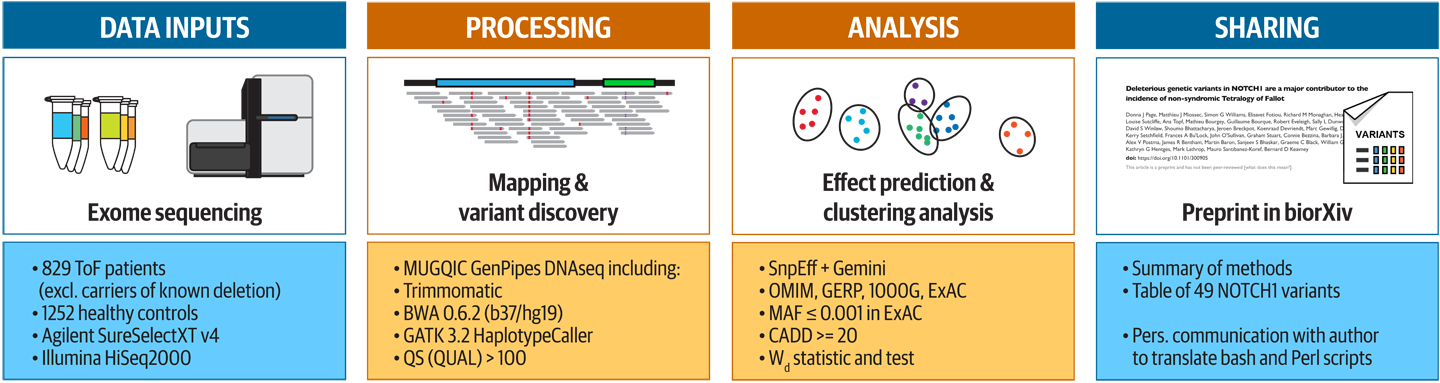
And that’s it; that’s our book in a nutshell. Four hundred pages summarized in a couple dozen sentences and images… Hopefully this gives you enough of a taste to help you gauge whether the book might be a good fit for you.
In the next installment of “What’s in the can?”, we’ll talk about the semi-official companion booklet we made for the book figures.
