CRISPResso and BEV notebook Sequence Orientations¶
Please note: All sequences are written in the 5' to 3' direction.
Inputs for running CRISPResso¶
Batch file (BEV input) columns:
| name | identifier | amplicon_seq | guide_seq | w | wc | exclude_bp_from_left | exclude_bp_from_right | plot_window_size |
|---|---|---|---|---|---|---|---|---|
This document walks through the amplicon_seq and guide_seq columns. For all other parameter descriptions, please see the Base Editor Validation Pipeline documentation on GitHub.
guide_seq
Since CRISPResso only uses the guide_seq input for naming files and does not use the guide_seq for alignment, for simplicity and consistency,
- the guide_seq should be written as designed, irrespective of primer direction
- guide_seq always goes 5’-3’ with PAM on 3’ end
- guide sense strand: the strand that contains the sequence that matches the mRNA

amplicon_seq
- must always be on the guide sense strand, irrespective of primer direction
Example:
![]()
Input for Validation Notebooks¶
Get translation_ref_seq¶
- Go to the Alleles_frequency_table_aroundsgRNA(sgRNA_sequence).txt file
- In the WT allele row (i.e. Unedited = TRUE), the Reference_Sequence is your CRISPResso reference sequence.

Compare CRISPResso reference sequence to translation reference sequence¶
The reference sequence that CRISPResso outputs is a certain number of nucleotides (determined by the quantification window parameter) upstream and downstream of the input guide sequence. Therefore, it will be in the guide sense direction.
Example:
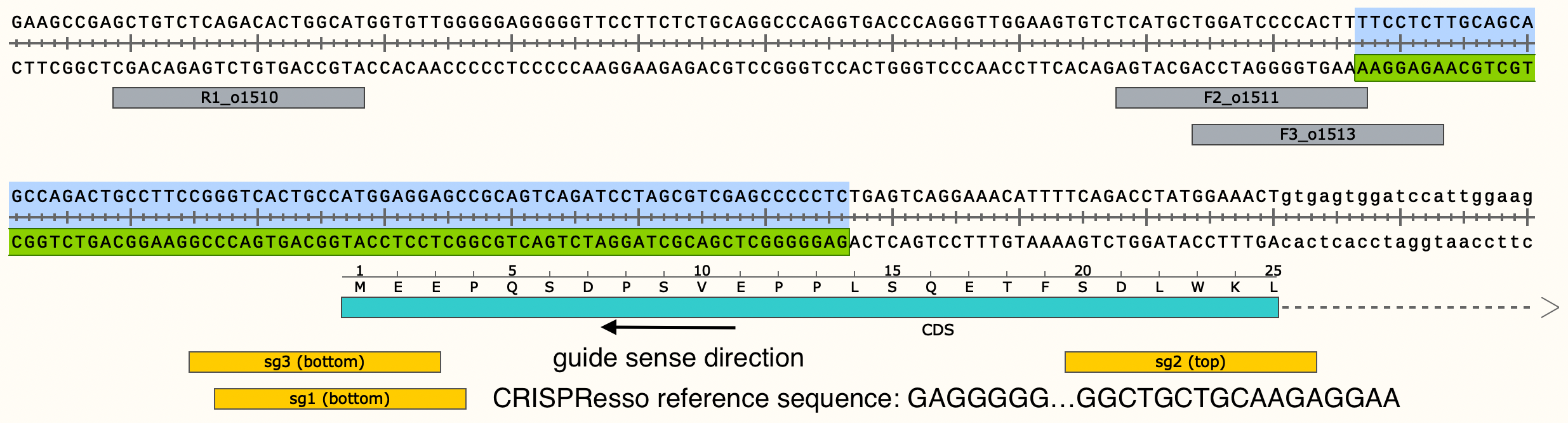
The sequence that should be translated to determine the amino acid sequence for a particular allele may not necessarily be the same as the CRISPResso reference sequence. The sequence that should be translated is what should be entered into the translation_ref_seq column of the metadata input file for the BEV_allele_frequencies validation notebook. This reference sequence should be formatted such that any untranslated regions (if applicable) are in lowercase.
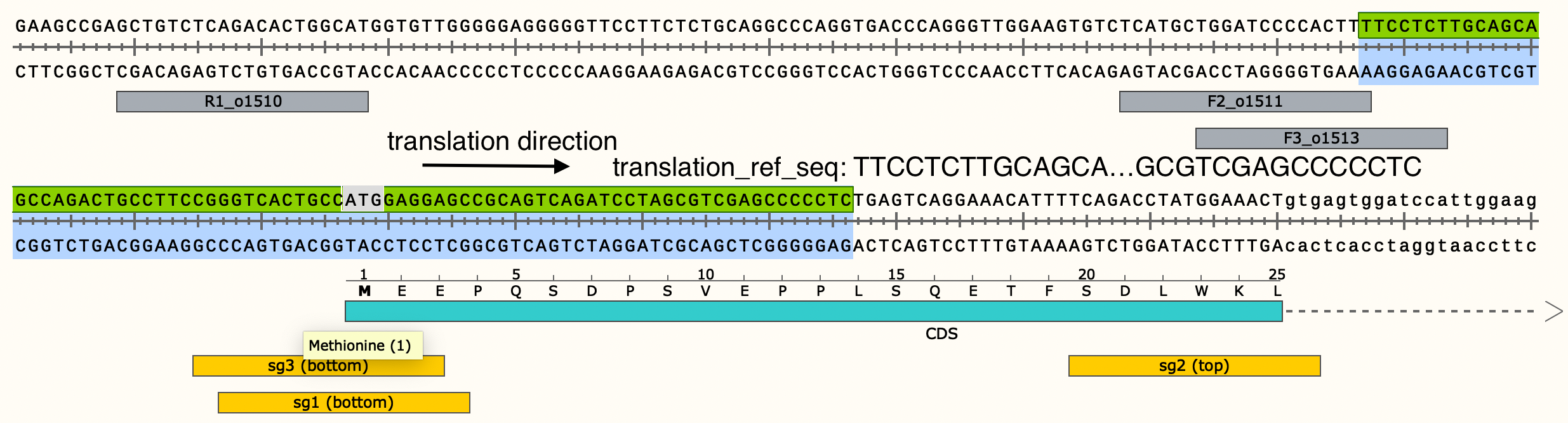
In this case, the forward DNA strand is being translated, so the reference sequence for translation is the reverse complement of the reference sequence that CRISPResso outputs. Therefore, in this case the reference sequence for the notebook metadata file is TTCCTCTTGCAGCAGCCAGACTGCCTTCCGGGTCACTGCCATGGAGGAGCCGCAGTCAGATCCTAGCGTCGAGCCCCCTC.
rev_com parameter¶
The rev_com parameter in the notebook input file determines which strand will be translated. The parameter is defined by the following:
- If the guide sequence/CRISPResso reference sequence and translation reference sequence are on opposite strands, then rev_com is True
- If the guide sequence/CRISPResso reference sequence and translation reference sequences are on the same strand, then rev_com is False
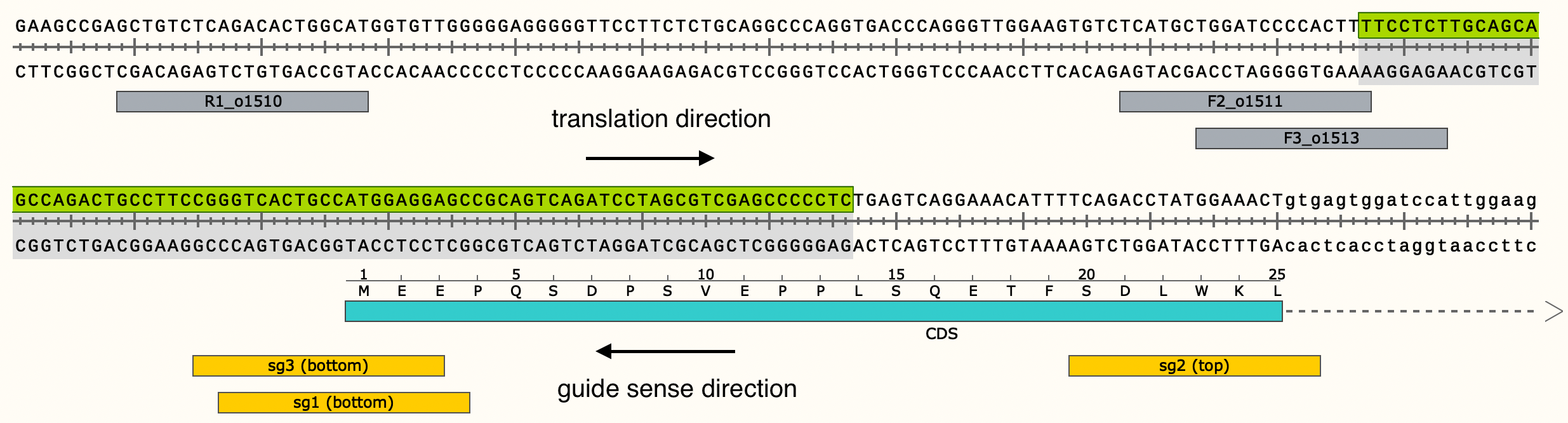
In this case, since the guide sequence and CRISPResso reference sequence are on the reverse DNA strand, while the strand being translated is the forward DNA strand, rev_com is True.
Here is what the metadata file for the BEV_allele_frequencies notebook would look like for this example. Explanations for the rest of the columns can be found in the BEV_allele_frequencies notebook:
| sg | sgRNA_sequence | translation_ref_seq | BEV_start | BEV_end | primer | frame | first_codon | last_codon | rev_com | BEV_ref | BEV_test |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GCTCCTCCATGGCAGTGACC | [TTCCTCTTGCAGCAGCCAGACTGCCTTCCGGGTCACTGCC]ATGGAGGAGCCGCAGTCAGATCCTAGCGTCGAGCCCCCTC | 417 | 426 | F3_R2 | 1 | ATG | CTG | True | 417;418 | 425;426 |